
also point out the necessity to consider interactions between sequence positions.
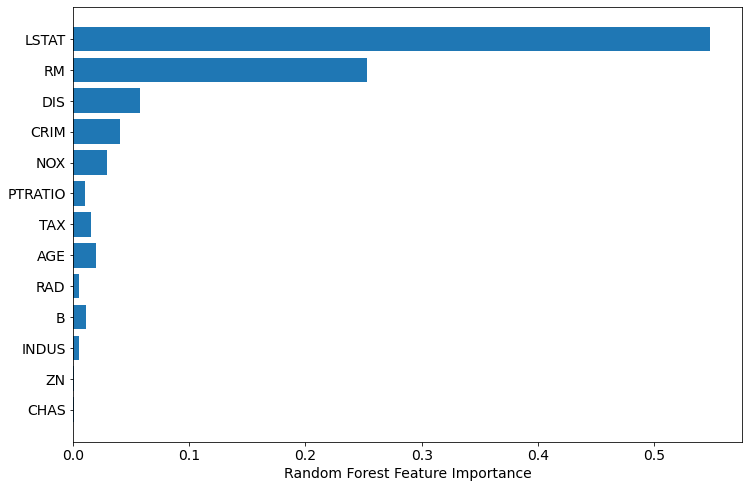
In the analysis of amino acid sequence data Segal et al. find that genetic markers relevant in interactions with other markers or environmental variables can be detected more efficiently by means of random forests than by means of univariate screening methods like Fisher's exact test. In this case a key advantage of random forest variable importance measures, as compared to univariate screening methods, is that they cover the impact of each predictor variable individually as well as in multivariate interactions with other predictor variables. By means of variable importance measures the candidate predictor variables can be compared with respect to their impact in predicting the response or even their causal effect (see, e.g., for assumptions necessary for interpreting the importance of a variable as a causal effect). Identifying relevant predictor variables, rather than only predicting the response by means of some "black-box" model, is of interest in many applications.

Recently, the variable importance measures yielded by random forests have also been suggested for the selection of relevant predictor variables in the analysis of microarray data, DNA sequencing and other applications. They show high predictive accuracy and are applicable even in high-dimensional problems with highly correlated variables, a situation which often occurs in bioinformatics. Within the past few years, random forests have become a popular and widely-used tool for non-parametric regression in many scientific areas.


 0 kommentar(er)
0 kommentar(er)
